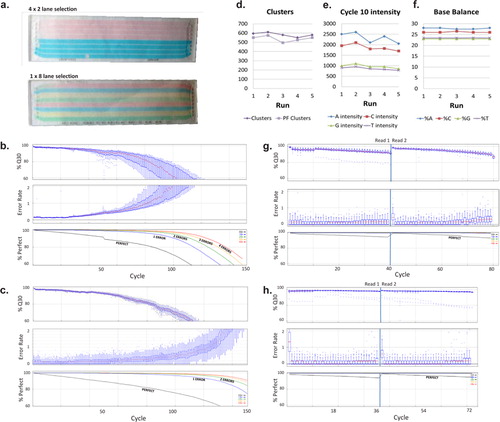
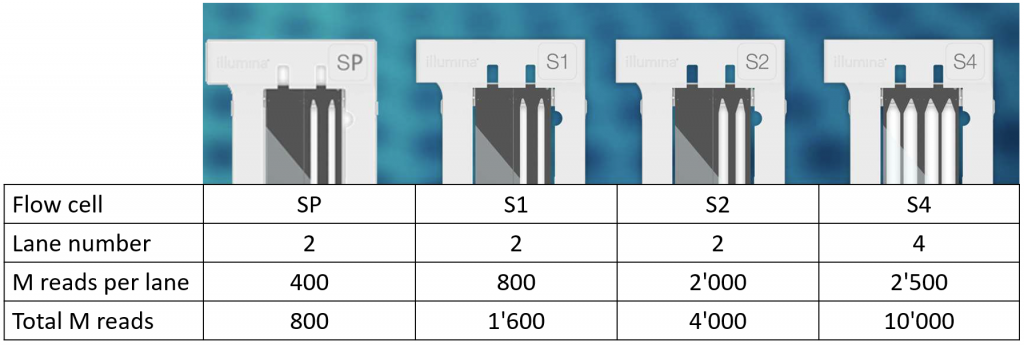
Quality score for each lane of Illumina reads. The line diagram of the... | Download Scientific Diagram

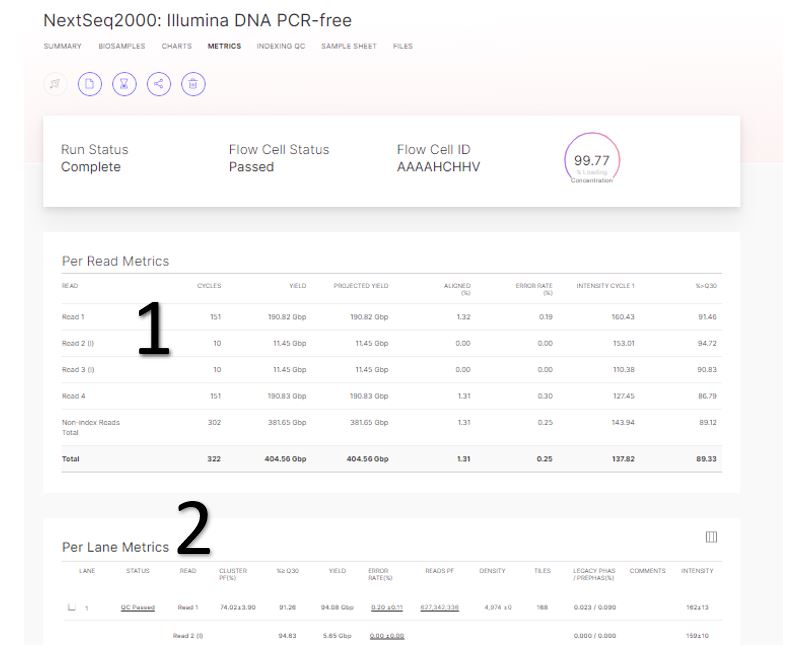
Generating FASTQs with cellranger-dna mkfastq -Software -Single Cell CNV -Official 10x Genomics Support

Quality score for each lane of Illumina reads. The line diagram of the... | Download Scientific Diagram

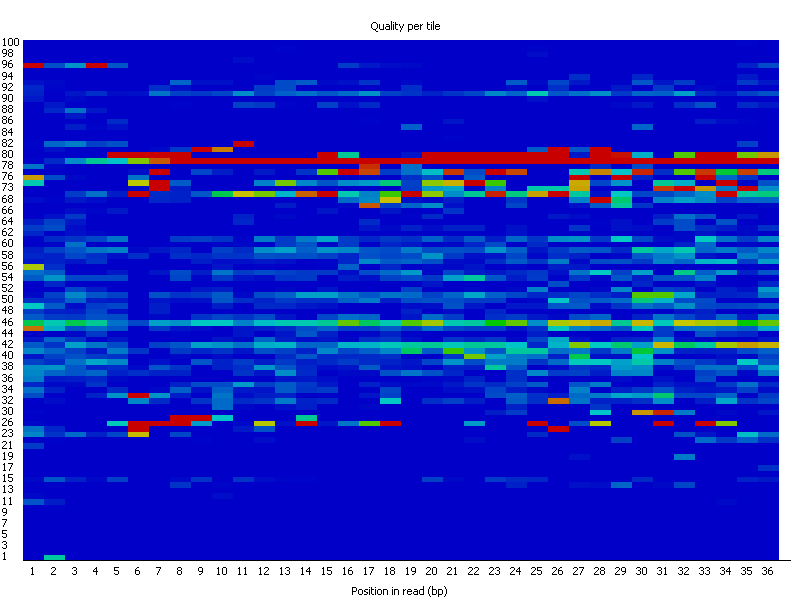
FASTQC output per sequencing lane. All four lanes 5–8, corresponding to... | Download Scientific Diagram
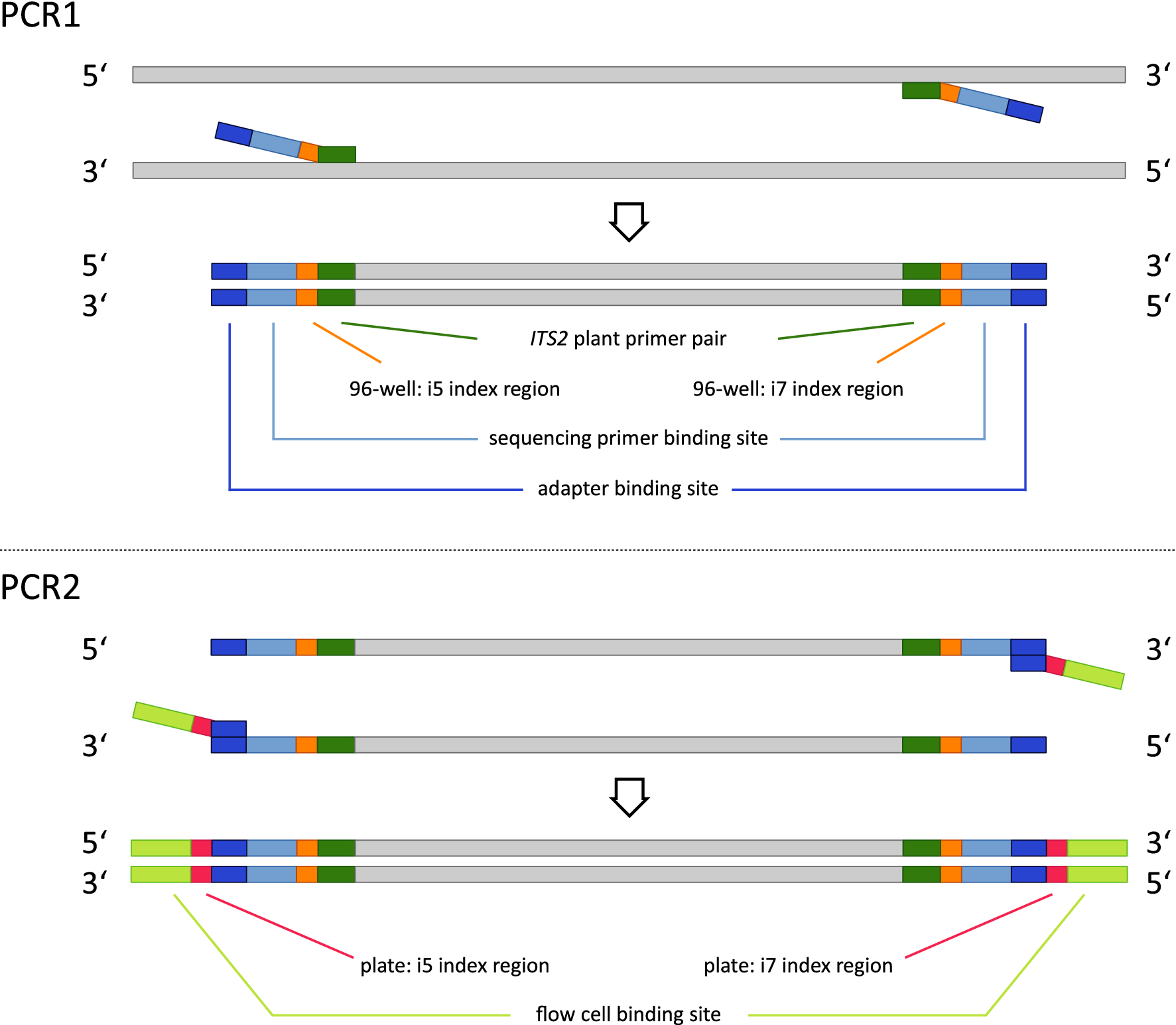
Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports

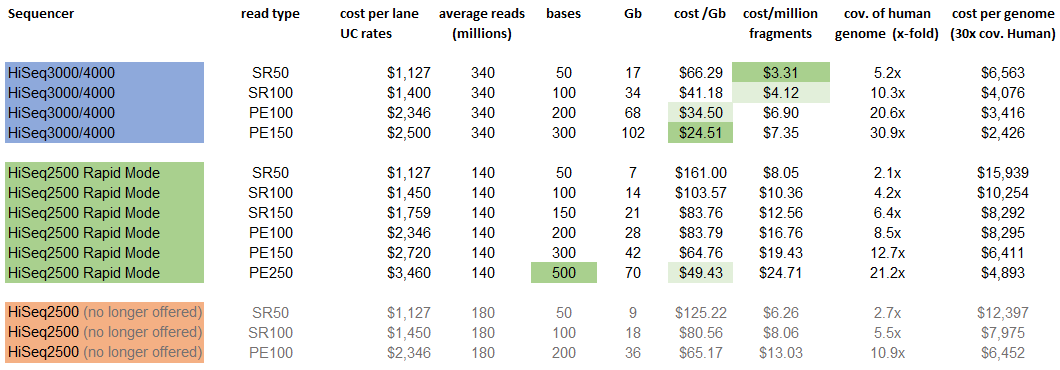
Raw Illumina sequence data files (for Dummies): Part 1 – What You Can Learn Just Looking at your Files – kscbioinformatics